Observation series
The MicroBrest observation series
Prokaryotic microorganisms (archaea and bacteria) are essential to the functioning of marine ecosystems, where they participate in the primary production, degradation and remineralisation of organic matter. Due to their abundance (several million in one litre of seawater) and diversity, these microorganisms play a central role in the biogeochemical cycles of the oceans. However, knowledge of their ecology remained very limited until the 2000s and the development of massive DNA sequencing technologies, making it possible to study them at the community level.
It is in this context that the MicroBrest observatory was developed. It is a genomic observatory of marine prokaryotic microorganisms in the Rade de Brest and Mer d’Iroise. It is mainly based on a time series initiated in 2014 at the SOMLIT station in Sainte-Anne du Portzic in order to finely characterise the seasonal dynamics of these microbial communities. Also, 4 campaigns conducted in the Iroise Sea at the Ouessant Front, as part of the LabexMer “M2BiPAT” project (2014-2015). These two monitoring programmes will allow us to better characterise the dynamics (seasonal and spatial) of bacteria and archaea, their functional role in the environment and their adaptation mechanisms in this coastal ecosystem.
Partners
The site
The sampling of the time series was carried out in the Goulet of the Brest harbour (48°21’32.17” N, 4°33’07.21” W), at the same time as that of the Somlit-Brest network, in order to benefit from the numerous parameters measured (physical, chemical and biological) by the Somlit network and thus to link the dynamics of the microbial communities with a precise environmental context.
The stations in the Iroise Sea were selected to cross the Ushant front, as determined by the physicists present during the M2BiPAT campaigns.
Monitoring
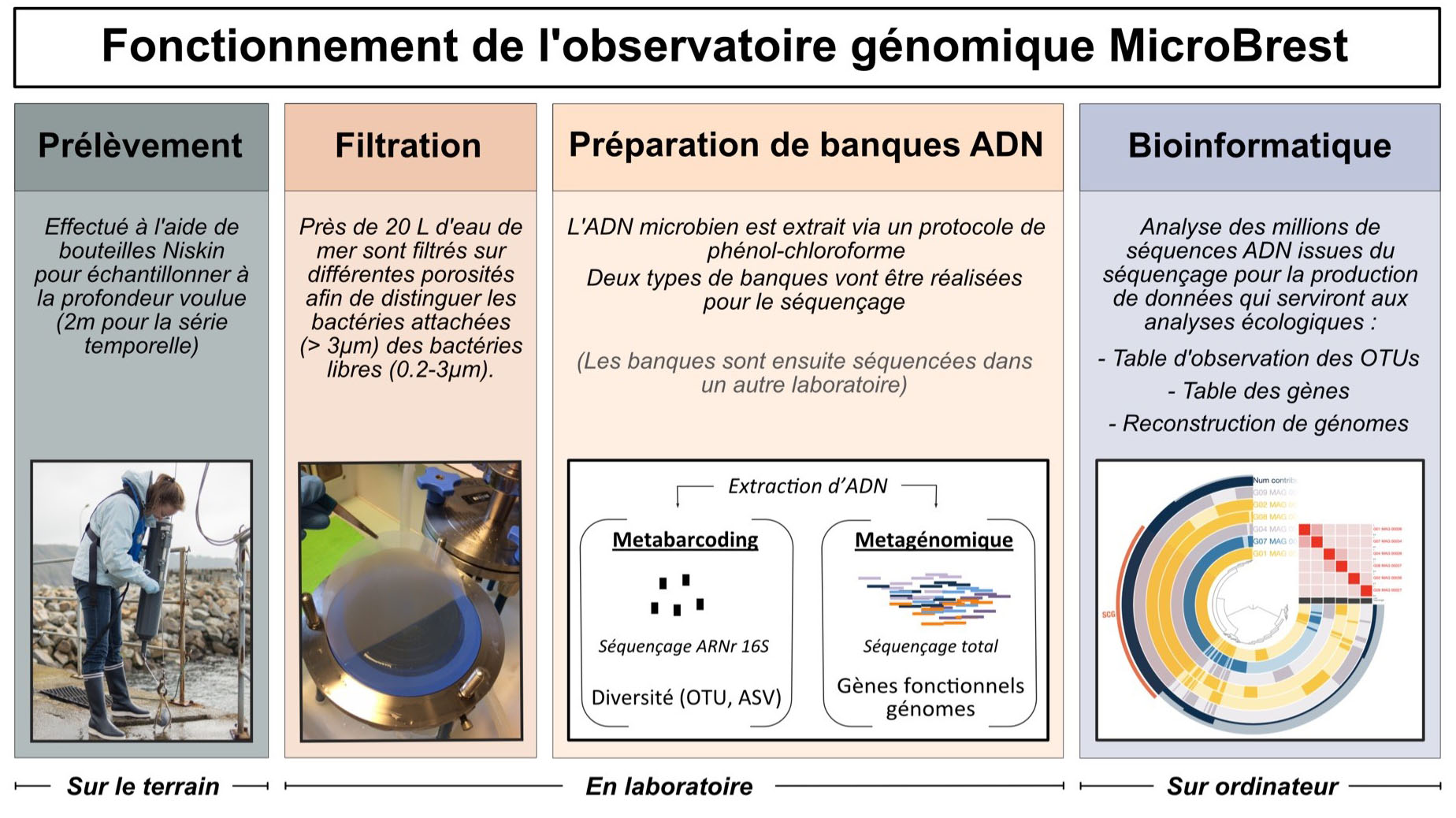
The series aims to characterise the communities of prokaryotes (bacteria and archaea) present in the water column. To do this, it uses two massive DNA sequencing approaches: metabarcoding and metagenomics. The first technology targets a marker gene, 16S rRNA, which provides information on the diversity of bacteria/archaea. The second technology sequences all the DNA contained in a sample, providing information on the functional genes present and even reconstructing bacterial/archaeal genomes, then called Metagenome-Assembled Genomes (or MAGs). In this network, a distinction is made between the free-living fraction of prokaryotes, (i.e. those that live freely in seawater) and the attached fraction (i.e. those that evolve attached to particles in the water column, or associated with phytoplankton, zooplankton for example).