Nitrogen fixation in the widely distributed marine gammaproteobacteria diazotroph Candidatus Thalassolituus haligoni
Abstract
The high diversity and global distribution of heterotrophic bacterial diazotrophs (HBDs) in the ocean has recently become apparent. However, understanding the role these largely uncultured microorganisms play in marine N2 fixation poses a challenge due to their undefined growth requirements and the complex regulation of the nitrogenase enzyme. We isolated and characterized Candidatus Thalassolituus haligoni, a member of a widely distributed clade of HBD belonging to the Oceanospirillales. Analysis of its nifH gene via amplicon sequencing revealed the extensive distribution of Cand. T. haligoni across the Pacific, Atlantic, and Arctic Oceans. Pangenome analysis indicates that the isolate shares >99% identity with an uncultured metagenome-assembled genome called Arc-Gamma-03, recently recovered from the Arctic Ocean. Through combined genomic, proteomic, and physiological approaches, we confirmed that the isolate fixes N2 gas. However, the mechanisms governing nitrogenase regulation in Cand. T. haligoni remain unclear. We propose Cand. T. haligoni as a globally distributed, cultured HBD model species within this understudied clade of Oceanospirillales.
Figure
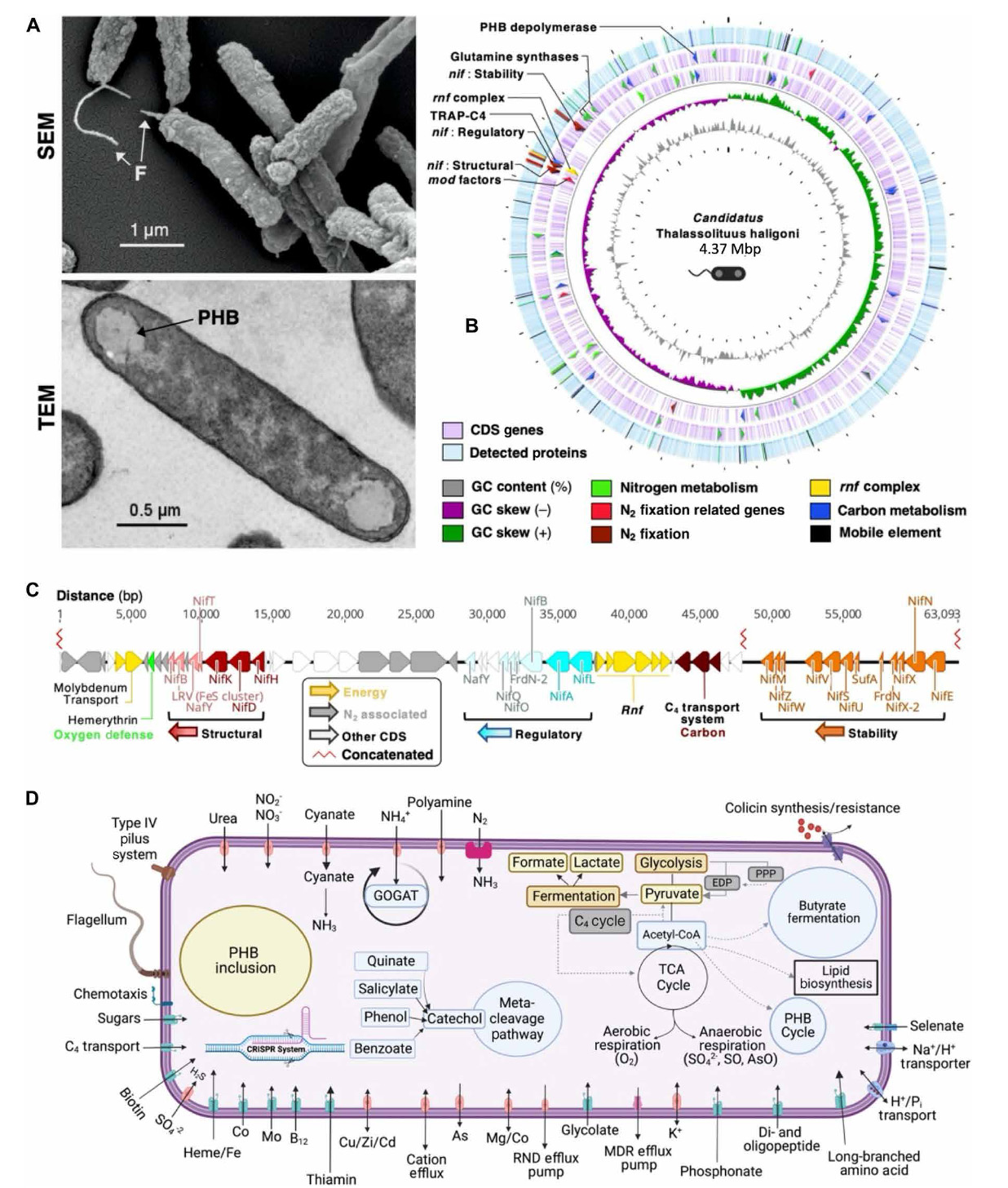
Fig. 1. Scanning electron microscopy and genomic, proteomic, and genome-assisted metabolic models of Cand. T. haligoni.
(A) (A) cell morphology of the isolate under n2- fixing conditions using scanning electron microscopy (SeM; top) and transmission electron microscopy (teM; bottom). SeM magnification = 17.71 × 103. Arrows indicate flagella (F) and PhB granules (PhB).
(B) CProteome-genome circular map under nO3− depleted conditions. Starting from the innermost ring: Gc content (gray), Gc skew for forward (green) and reverse strands (magenta), leading and lagging strand cdSs (light purple), and detected proteins (blue). Arrows in the genome indicate genes of interest [nif clusters, n2 fixation–related genes are molybdate uptake and transformation genes (mod factors), tRAP-c4 uptake, and glutamine synthesis] and have corresponding detected proteins highlighted in respective colors. White spaces in the proteome indicate regions not detected.
(C) Nif clusters (red, blue, and orange) and associated n2 fixation genes (gray). n2 fixation–associated genes include glycogen phosphorylase and glycogen uptake genes. numbers along the line indicate base-pair locations within the genome. A nucleotide break in the center of the diagram from 3,598,007 nt to 3,737,467 nt is represented by the zig-zag line in red (concatenated). Clusters are color-coded to the associated nif clusters, with accessory genes to that cluster indicated by respective color hues.
(D) Simplified metabolic schematic of the isolate based on the genome annotation. the schematic was created using the RASt server and BioRender.
Reference
e. “Nitrogen fixation in the widely distributed marine γ-proteobacterial diazotroph Candidatus Thalassolituus haligoni“. Science Advances Volume 10, Issue 31 (Aug 2024)