Spatial distribution of tropical fish assemblages
Comprehensive spatial distribution of tropical fish assemblages from multifrequency acoustics and video fulfils the island mass effect framework.
Describing fish distribution and associated environmental features is the first step toward understanding how fish communities are spatially structured and is a necessary step to conduct Marine Spatial Planning (MSP) and operate relevant protection policies.
Abstract
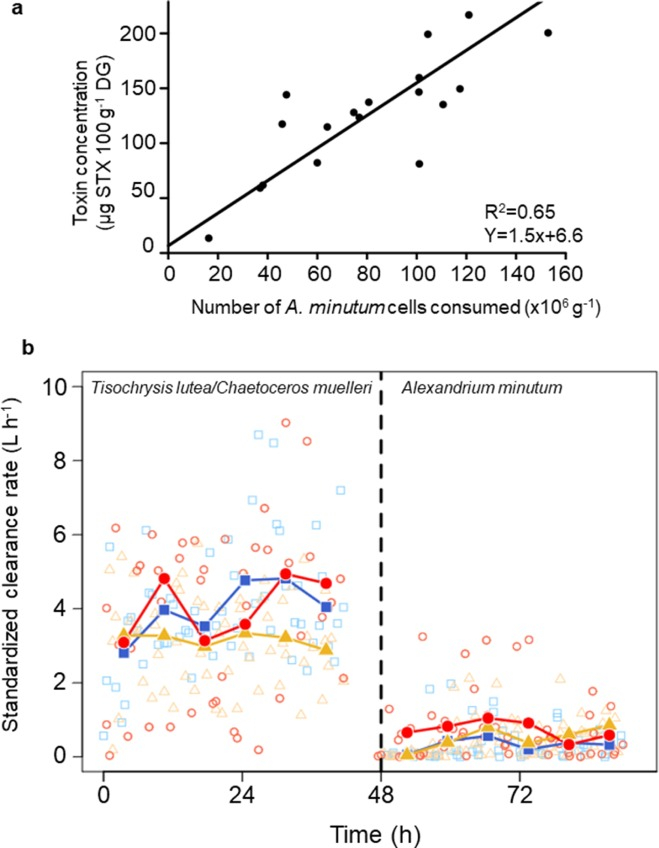
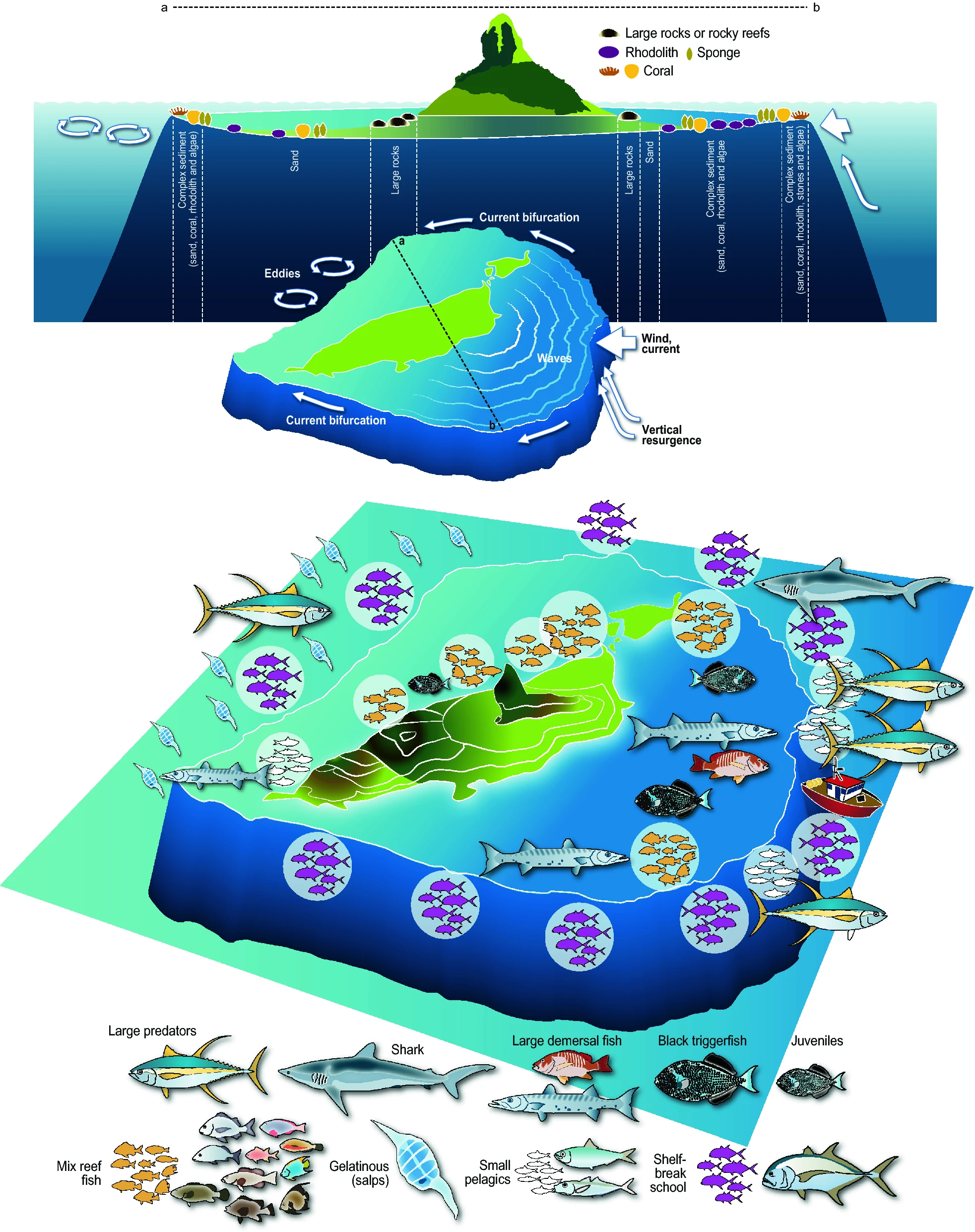
Tropical marine ecosystems are highly biodiverse and provide resources for small-scale fisheries and tourism. However, precise information on fish spatial distribution is lacking, which limits our ability to reconcile exploitation and conservation. We combined acoustics to video observations to provide a comprehensive description of fish distribution in a typical tropical environment, the Fernando de Noronha Archipelago (FNA) off Northeast Brazil. We identified and classified all acoustic echoes into ten fish assemblage and two triggerfish species. This opened up the possibility to relate the different spatial patterns to a series of environmental factors and the level of protection. We provide the first biomass estimation of the black triggerfish Melichthys niger, a key tropical player. By comparing the effects of euphotic and mesophotic reefs we show that more than the depth, the most important feature is the topography with the shelf-break as the most important hotspot. We also complete the portrait of the island mass effect revealing a clear spatial dissymmetry regarding fish distribution. Indeed, while primary productivity is higher downstream, fish concentrate upstream. The comprehensive fish distribution provided by our approach is directly usable to implement scientific-grounded Marine Spatial Planning..
Synthetic representation of the island mass effect as illustrated by the case of Fernando de Noronha.


 (c) Gert Hansen - Licensed under a Creative Commons Attribution-Noncommercial-Share Alike 4.0 License
(c) Gert Hansen - Licensed under a Creative Commons Attribution-Noncommercial-Share Alike 4.0 License