In Silico Analysis of Pacific Oyster (Crassostrea gigas) Transcriptome over Developmental Stages Reveals Candidate Genes for Larval Settlement
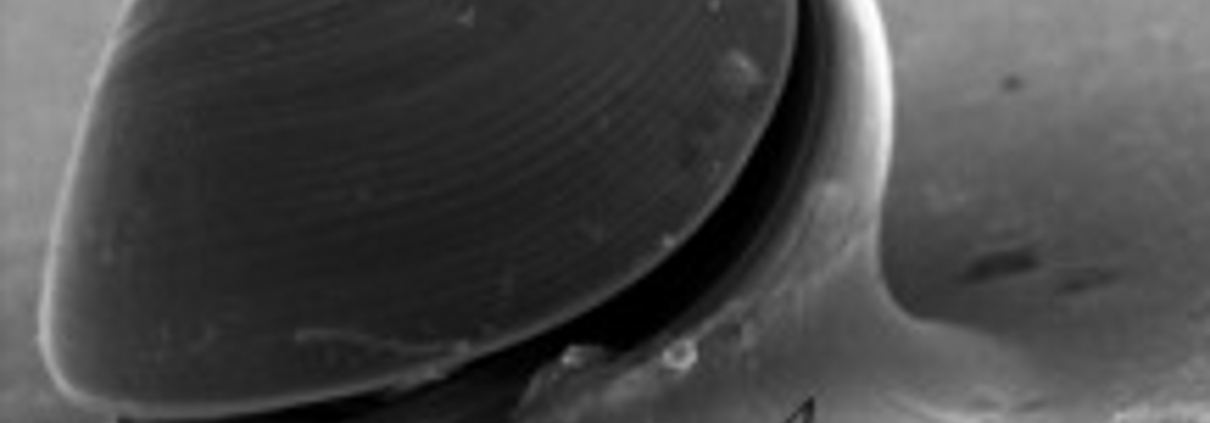
Abstract: Following their planktonic phase, the larvae of benthic marine organisms must locate a suitable habitat to settle and metamorphose. For oysters, larval adhesion occurs at the pediveliger stage with the secretion of a proteinaceous bioadhesive produced by the foot, a specialized and ephemeral organ. Oyster bioadhesive is highly resistant to proteomic extraction and is only produced in very low quantities, which explains why it has been very little examined in larvae to date. In silico analysis of nucleic acid databases could help to identify genes of interest implicated in settlement. In this work, the publicly available transcriptome of Pacific oyster Crassostrea gigas over its developmental stages was mined to select genes highly expressed at the pediveliger stage. Our analysis revealed 59 sequences potentially implicated in adhesion of C. gigas larvae. Some related proteins contain conserved domains already described in other bioadhesives. We propose a hypothetic composition of C. gigas bioadhesive in which the protein constituent is probably composed of collagen and the von Willebrand Factor domain could play a role in adhesive cohesion. Genes coding for enzymes implicated in DOPA chemistry were also detected, indicating that this modification is also potentially present in the adhesive of pediveliger larvae.
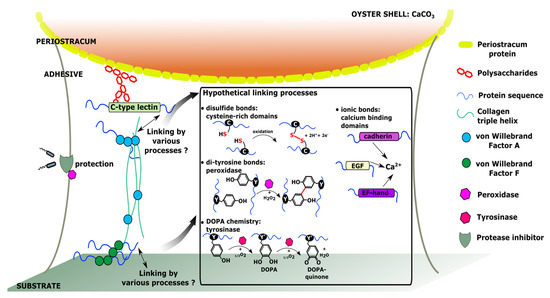
Schematic representation of the hypothetical molecular interactions involved in the adhesion of C. gigas pediveliger larvae, based on the selection of genes specifically expressed at the pediveliger stage.
REFERENCE :
The majority of bioadhesives secreted by animals are composed of proteins that allow permanent or reversible links to the substrate. In the marine environment, these glues are efficient in wet conditions and could thus potentially represent useful alternatives to synthetic adhesives, particularly for biomedical applications.