Mercury isotope clocks predict coastal residency and migration timing of hammerhead sharks
Abstract and highlights
- The management of migratory taxa relies on the knowledge of their movements. Among them, ontogenetic habitat shift, from nurseries to adult habitats, is a behavioural trait shared across marine taxa allowing resource partitioning between life stages and reducing predation risk. As this movement is consistent over time, characterizing its timing is critical to implement efficient management plans, notably in coastal areas to mitigate the impact of fisheries on juvenile stocks.
- In the Mexican Pacific, habitat use of the smooth hammerhead shark (Sphyrna zygaena) is poorly described, while the species is heavily harvested. Given the large uncertainties associated with the timing of out-migration from coastal nursery grounds to offshore waters prior to reproductive maturity, a more precise assessment of smooth hammerhead shark movements is needed.
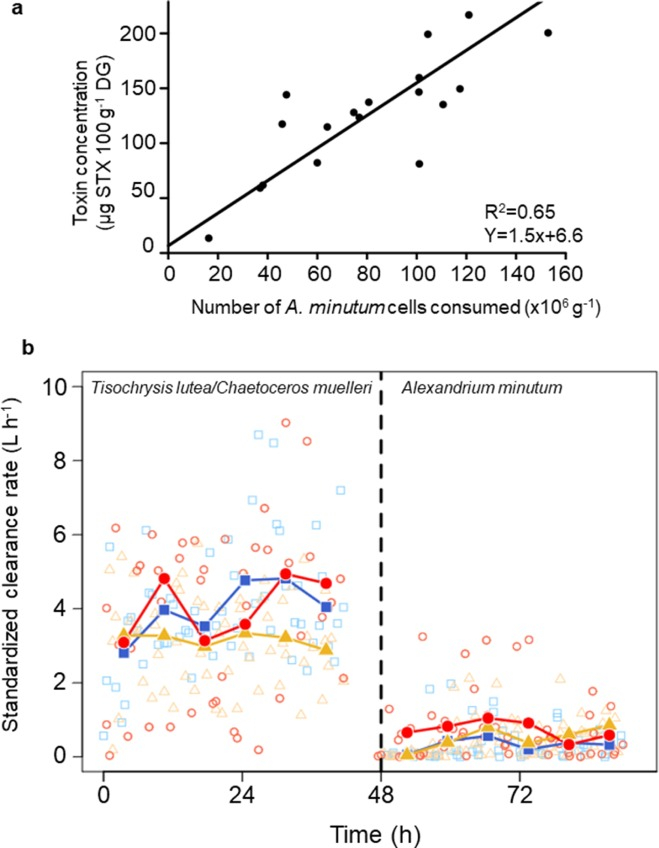
- Photochemical degradation of mercury imparts mass-independent isotope fractionation (Δ199Hg) which can be used to discriminate between neonate coastal shallow habitats and the offshore deep foraging patterns of late juveniles. Here, we present the application of muscle Δ199Hg as molecular clocks to predict the timing of ontogenetic habitat shifts by smooth hammerhead sharks, based on their isotopic compositions at the initial and arrival habitats and on muscle isotopic turnover rate.
- We observed decreases in Δ199Hg values with shark body length, reflecting increasing reliance on offshore mesopelagic prey with age. Coastal residency estimates indicated that smooth hammerhead sharks utilize coastal resources for 2 years prior to offshore migration, suggesting a prolonged residency in these ecosystems.
- Policy implications. This study demonstrates how mercury stable isotopes and isotopic clocks can be implemented as a complementary tool for stock management by predicting the timing of animal migration—a key aspect in the conservation of marine taxa. In the Mexican Pacific, fishing pressure on shark species occurs in coastal habitats depleting juvenile stocks. Consequently, management decision support tools are imperative for effectively maintaining early life stage population levels over time. The finding that smooth hammerhead sharks extensively rely on highly fished habitats for 2 years after parturition supports the relevance of establishing a size limit in coastal fisheries and demonstrates how the current temporal shark fishing closure could lack efficiency for the species.
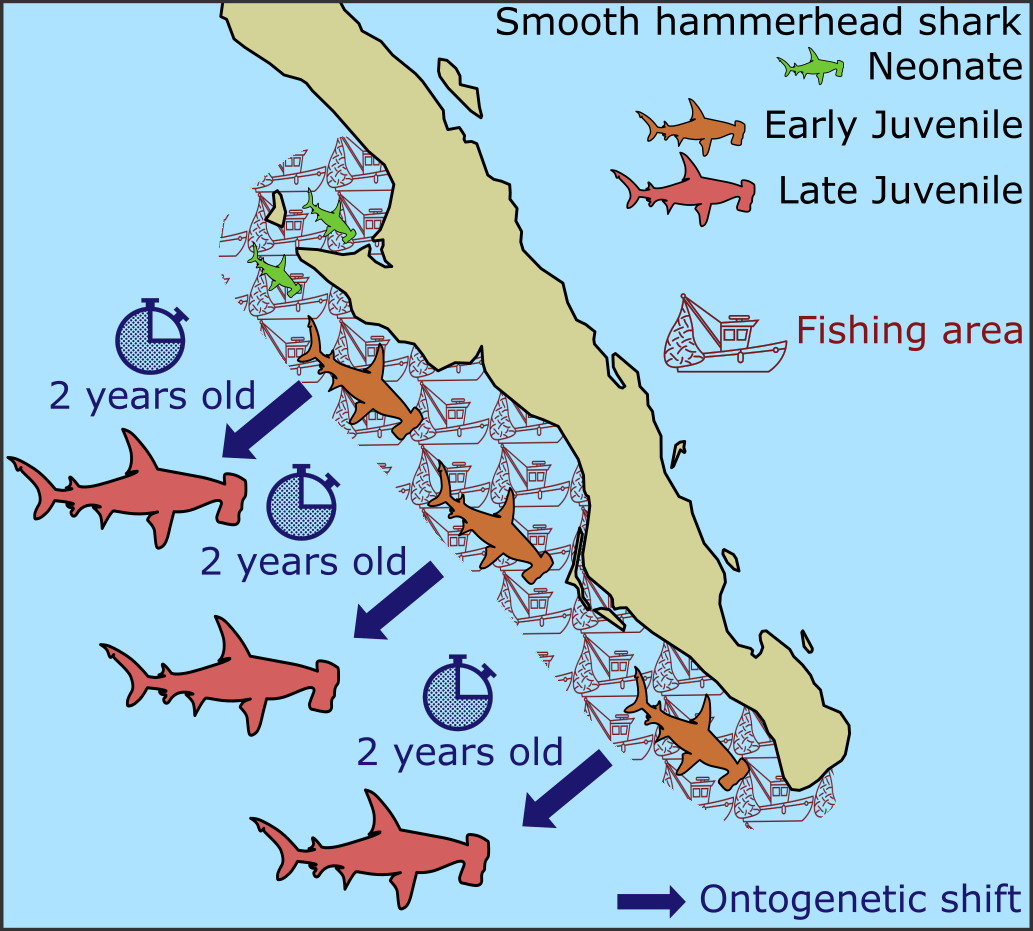
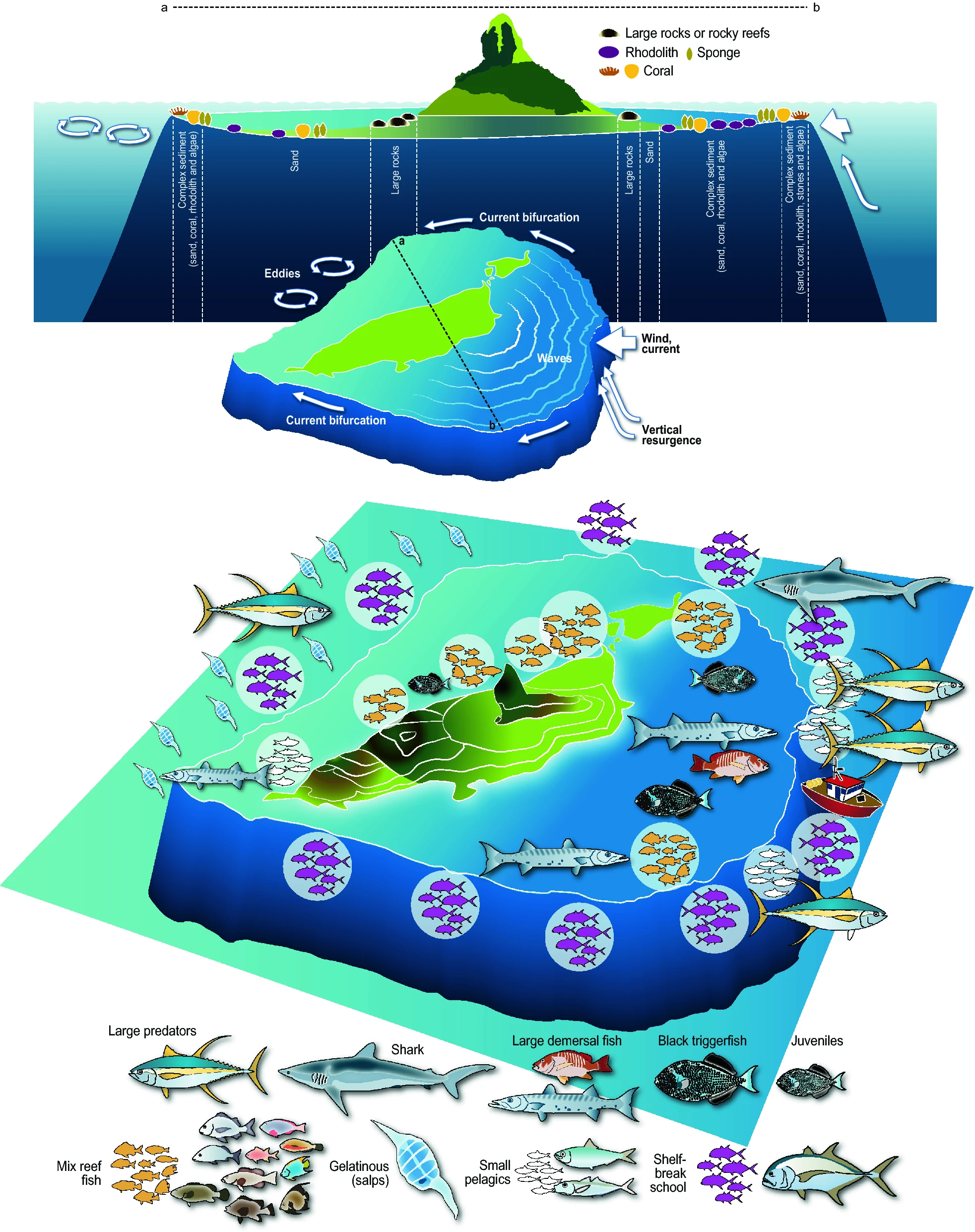
Graphical abstract.
Reference
Besnard, L., Lucca, B. M., Shipley, O. N., Le Croizier, G., Martínez-Rincón, R. O., Sonke, J. E., Point, D., Galván-Magaña, F., Kraffe, E., Kwon, S. Y., & Schaal, G. (2023). Mercury isotope clocks predict coastal residency and migration timing of hammerhead sharks. Journal of Applied Ecology, 60, 803–813. https://doi.org/10.1111/1365-2664.1438





 (c) Gert Hansen - Licensed under a Creative Commons Attribution-Noncommercial-Share Alike 4.0 License
(c) Gert Hansen - Licensed under a Creative Commons Attribution-Noncommercial-Share Alike 4.0 License